Dynamics of Respiratory Infections

Our research
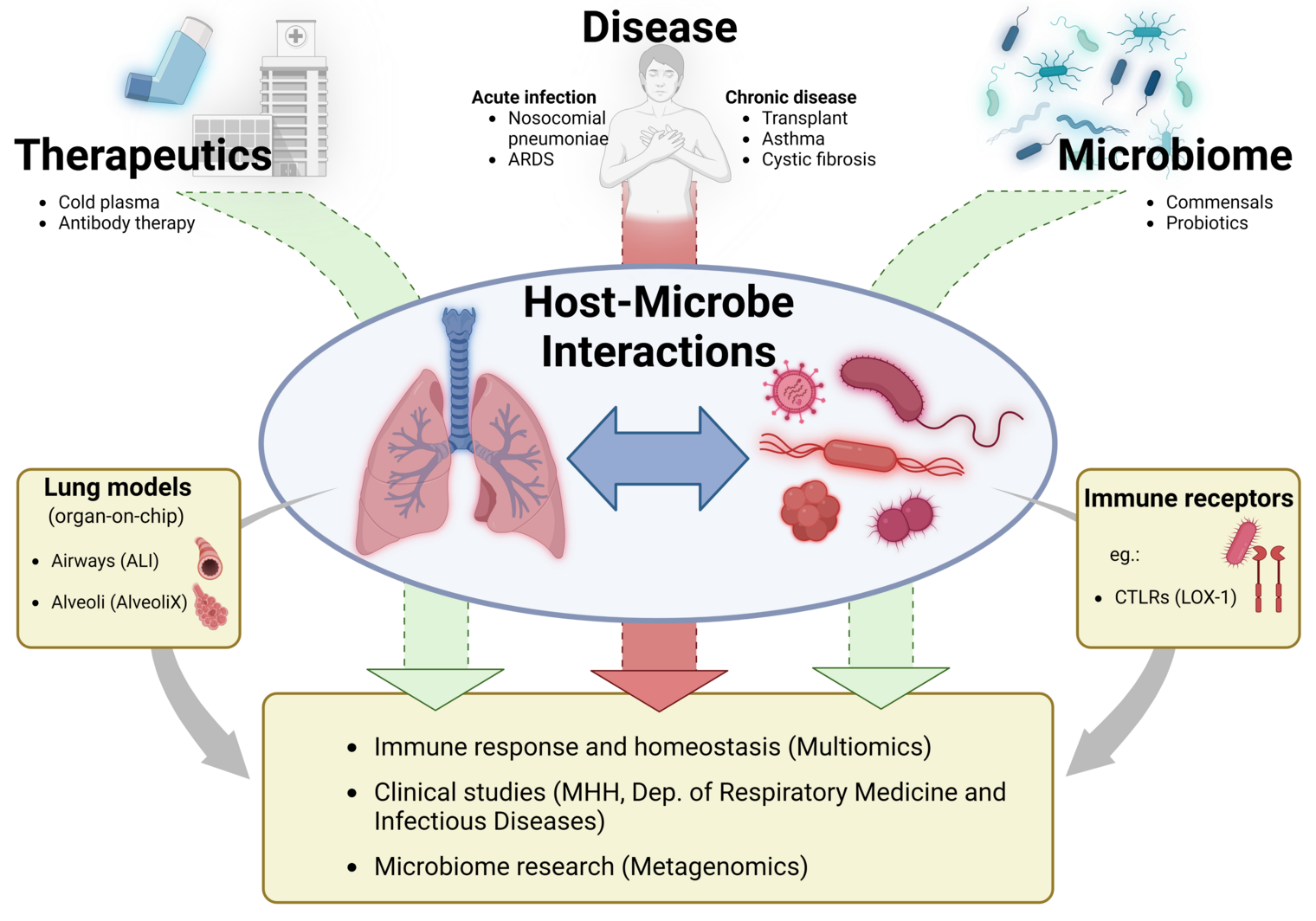
Microbiome, immune response and pathogenesis in the lung
The respiratory microbiome is a central, but as yet poorly understood, component of lung homeostasis. It contributes significantly to the maintenance of the basic immunological tone, to colonization resistance against pathogens and to the control of inflammatory processes. In contrast to the gut microbiome, however, the microbial communities of the lower respiratory tract have hardly been characterized to date - especially with regard to their functional role in health and disease.
A major reason for this lies in the methodological challenges: The microbial biomass in the lungs is low and the samples - e.g. from bronchoalveolar lavage - contain a high proportion of human DNA. In addition, there is a lack of standardized methods for the reliable analysis of non-bacterial microbes such as fungi (mycobiome) and viruses (virome). To date, there is a lack of comparative studies on the performance of different sequencing approaches, primer specificities and protocols for the analysis of microbial signatures from the lung.
Our research group is uniquely positioned at the interface of clinical infection medicine and translational microbiome research - as part of the Clinic for Pneumology and Infectiology at Hannover Medical School (MHH) and at the same time as the research group “Dynamics of Respiratory Infections (DINF)” at the Helmholtz Centre for Infection Research (HZI) in Braunschweig.
This dual anchoring enables direct feedback between clinical observation and basic scientific analysis. This structure allows us to transfer patient material and clinical questions into experimental systems in a targeted manner - and, conversely, to reflect molecular findings directly back into diagnostic or therapeutic applications. This makes it an ideal model for translational infection research with a focus on lung infections from the clinic to the laboratory and back.
We use state-of-the-art sequencing technologies - including amplicon-based ITS and 16S sequencing, shotgun metagenomics and long-read technologies - to enable the most comprehensive coverage of microbial communities possible. In addition, we develop computational evaluation strategies to reliably identify microbial signatures even with low biomass and a high proportion of human DNA. A particular focus is on the methodological development of the fungal components (mycobiome) of the respiratory microbiome, whose functional role in microbial networks and in the interaction with human cells is still largely unknown.
Our aim is to comprehensively characterize the microbial ecosystems of the lung and to better understand their role in the pathogenesis of respiratory diseases, such as community-acquired or nosocomial pneumonia. The focus is on classical pathogens such as Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae and Escherichia coli, as well as less studied commensals such as Streptococcus mitis, Prevotella spp., Moraxella catarrhalis and Haemophilus influenzae, which potentially exert immunomodulatory functions. In addition, we analyze the functional integration of fungal species - in particular Candida spp. and Aspergillus spp. - into the microbial communities of the lung and their interactions with bacterial microbes and host cells.
For functional analysis, we use advanced experimental platforms, including air-liquid interface (ALI) cultures, lung-on-chip systems, patient-derived lung tissue slices (PCLS) and human lung organoids. These models allow us to study the interactions between microorganisms, respiratory epithelium and alveolar macrophages in an immunologically relevant context.
In the long term, our research aims not only to better understand the microbial signature and dynamics in the lung, but also to harness them for clinical diagnostics and new therapeutic strategies - for example through targeted microbiome regulation, the identification of diagnostic markers or innovative approaches to the prevention and treatment of respiratory infections.
Our research
Microbiome, immune response and pathogenesis in the lung
The respiratory microbiome is a central, but as yet poorly understood, component of lung homeostasis. It contributes significantly to the maintenance of the basic immunological tone, to colonization resistance against pathogens and to the control of inflammatory processes. In contrast to the gut microbiome, however, the microbial communities of the lower respiratory tract have hardly been characterized to date - especially with regard to their functional role in health and disease.
A major reason for this lies in the methodological challenges: The microbial biomass in the lungs is low and the samples - e.g. from bronchoalveolar lavage - contain a high proportion of human DNA. In addition, there is a lack of standardized methods for the reliable analysis of non-bacterial microbes such as fungi (mycobiome) and viruses (virome). To date, there is a lack of comparative studies on the performance of different sequencing approaches, primer specificities and protocols for the analysis of microbial signatures from the lung.
Our research group is uniquely positioned at the interface of clinical infection medicine and translational microbiome research - as part of the Clinic for Pneumology and Infectiology at Hannover Medical School (MHH) and at the same time as the research group “Dynamics of Respiratory Infections (DINF)” at the Helmholtz Centre for Infection Research (HZI) in Braunschweig.
This dual anchoring enables direct feedback between clinical observation and basic scientific analysis. This structure allows us to transfer patient material and clinical questions into experimental systems in a targeted manner - and, conversely, to reflect molecular findings directly back into diagnostic or therapeutic applications. This makes it an ideal model for translational infection research with a focus on lung infections from the clinic to the laboratory and back.
We use state-of-the-art sequencing technologies - including amplicon-based ITS and 16S sequencing, shotgun metagenomics and long-read technologies - to enable the most comprehensive coverage of microbial communities possible. In addition, we develop computational evaluation strategies to reliably identify microbial signatures even with low biomass and a high proportion of human DNA. A particular focus is on the methodological development of the fungal components (mycobiome) of the respiratory microbiome, whose functional role in microbial networks and in the interaction with human cells is still largely unknown.
Our aim is to comprehensively characterize the microbial ecosystems of the lung and to better understand their role in the pathogenesis of respiratory diseases, such as community-acquired or nosocomial pneumonia. The focus is on classical pathogens such as Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae and Escherichia coli, as well as less studied commensals such as Streptococcus mitis, Prevotella spp., Moraxella catarrhalis and Haemophilus influenzae, which potentially exert immunomodulatory functions. In addition, we analyze the functional integration of fungal species - in particular Candida spp. and Aspergillus spp. - into the microbial communities of the lung and their interactions with bacterial microbes and host cells.
For functional analysis, we use advanced experimental platforms, including air-liquid interface (ALI) cultures, lung-on-chip systems, patient-derived lung tissue slices (PCLS) and human lung organoids. These models allow us to study the interactions between microorganisms, respiratory epithelium and alveolar macrophages in an immunologically relevant context.
In the long term, our research aims not only to better understand the microbial signature and dynamics in the lung, but also to harness them for clinical diagnostics and new therapeutic strategies - for example through targeted microbiome regulation, the identification of diagnostic markers or innovative approaches to the prevention and treatment of respiratory infections.
Prof Dr med Hortense Slevogt
A better understanding of the host-microbe interactions in the lung may help us to develop novel therapeutic strategies for a variety of respiratory infections.

Prof. Dr. med. Hortense Slevogt is an internist and infectiologist. She has been lead senior physician for Clinical Infectiology at the Clinic for Pneumology and Infectiology at Hannover Medical School (MHH) since 2022.
She has established herself as a distinguished researcher and physician in the fields of respiratory infections and immunology. Her academic journey began with a degree in medicine at Freie Universität Berlin, during which she developed a particular interest in infectious diseases and their impact on respiratory health. Driven by her passion for understanding the intricate workings of the human immune system, she pursued further specialization in Internal Medicine and Pulmonology, becoming a recognized expert in the field.
In 2009, Prof Slevogt's achievements were acknowledged through the completion of her habilitation in Internal Medicine at Charité - Universitätsmedizin Berlin. This significant milestone marked her expertise and profound understanding of the intricate nature of severe infections. A few years later, in 2011, Prof. Slevogt assumed the leadership of the Host Septomics Group at Universitätsklinikum Jena.
In 2022, she obtained the W3-Professorship at Medizinische Hochschule Hannover (MHH) and became the Head of the Research Group "Respiratory Infection Dynamics" (DINF) at the Helmholtz Centre for Infection Research (HZI) in Braunschweig. Her role entails spearheading translational projects that involve collaborations with esteemed academic and medical partners at the MHH. Through these endeavors, Dr Slevogt investigates host-pathogen interactomes and elucidates the mechanisms underlying immune responses and their control.
In parallel to her research pursuits, Dr Slevogt remains dedicated to clinical practice, ensuring that her findings directly benefit patients. She serves as the Senior Consultant in Clinical Infectiology at the Department of Respiratory Medicine and Infectious Diseases (MHH), contributing her expertise to the diagnosis and treatment of respiratory infections.
Team










Best Talk Award in Microbiology for Sarah Truthe at ZIB Retreat 2025
We are delighted to share that our PhD student Sarah Truthe (in the center) was awarded the Best Talk Award in Microbiology at the ZIB Program Retreat in June 2025. Her presentation convinced the audience, who voted her talk as the best in the microbiology section. Congratulations to Sarah on this well-deserved recognition of her excellent work!
Selected Publications
Zubiria-Barrera C), Yamba LY) , Klassert TE, Bos M, Ahl J, Wasserstrom L, Slevogt H,* Riesbeck K* (2025). Profiling the nasopharyngeal Microbiome in patients with community-acquired pneumonia caused by Streptococcus pneumoniae: diagnostic challenges and ecological insights. Med Microbiol Immunol. 2025 Apr 10;214(1):19. doi: 10.1007/s00430-025-00828-0.
Klassert TE, Zubiria-Barrera C, Denkel L, Neubert R, Schneegans A, Kulle A, Vester A, Bloos F, Schulze C, Epstude J, Gastmeier P, Geffers C, Slevogt H (2024). Skin dysbiosis and loss of microbiome site specificity in critically ill patients. Microbiol Spectr. 2024 Mar 5;12(3):e0307823. doi: 10.1128/spectrum.03078-23.
Müller MM, Baldauf C, Hornischer S, Klassert TE, Schneegans A, Behnert A, Pletz MW, Hagel S, Slevogt H §. (2023). Staphylococcus aureus induces tolerance in human monocytes accompanied with expression changes of cell surface markers. Front Immunol. 14:1046374. doi: 10.3389/fimmu.2023.1046374.
Klassert TE , Leistner R, Zubiria-Barrera C, Stock M, López M, Neubert R, Driesch D, Gastmeier P, Slevogt H §. (2021). Bacterial colonization dynamics and antibiotic resistance gene dissemination in the hospital environment after first patient occupancy: a longitudinal metagenetic study. Microbiome. 9(1):169. doi: 10.1186/s40168-021-01109-7.
Lehmann R, Müller MM, Klassert TE, Driesch D, Stock M, Heinrich A, Conrad T, Moore C, Schier UK, Guthke R, Slevogt H§. (2018). Differential regulation of the transcriptomic and secretomic landscape of sensor and effector functions of human airway epithelial cells. Mucosal Immunol. 11(3):627-642. doi: 10.1038/mi.2017.100.
Projects
- The influence of microbial commensal-pathogen interactions on shaping the immunological environment of the human respiratory tract (RESIST)
- PlasmaCare: Analysis of the safety of cold plasma application on human lung epithelium using complex cell culture models.
- The interplay of Candida albicans and Aspergillus fumigatus induced signals in receptor-mediated modulation of antifungal immune responses
- Development of a novel test kit for the diagnosis of pneumocystis jirovecii infection