Molecular Structural Biology

Our research
The Department of Molecular Structural Biology is focussing on highresolution structural elucidation of large macromolecular complexes and machines that are important for infection processes. State-of-the-art methods such as single-particle cryo-electron microscopy (cryo-EM) and X-ray crystallography are used for this purpose. The structural biology analyses are complemented by a broad spectrum of molecular biological, biochemical and bio-physical methods.
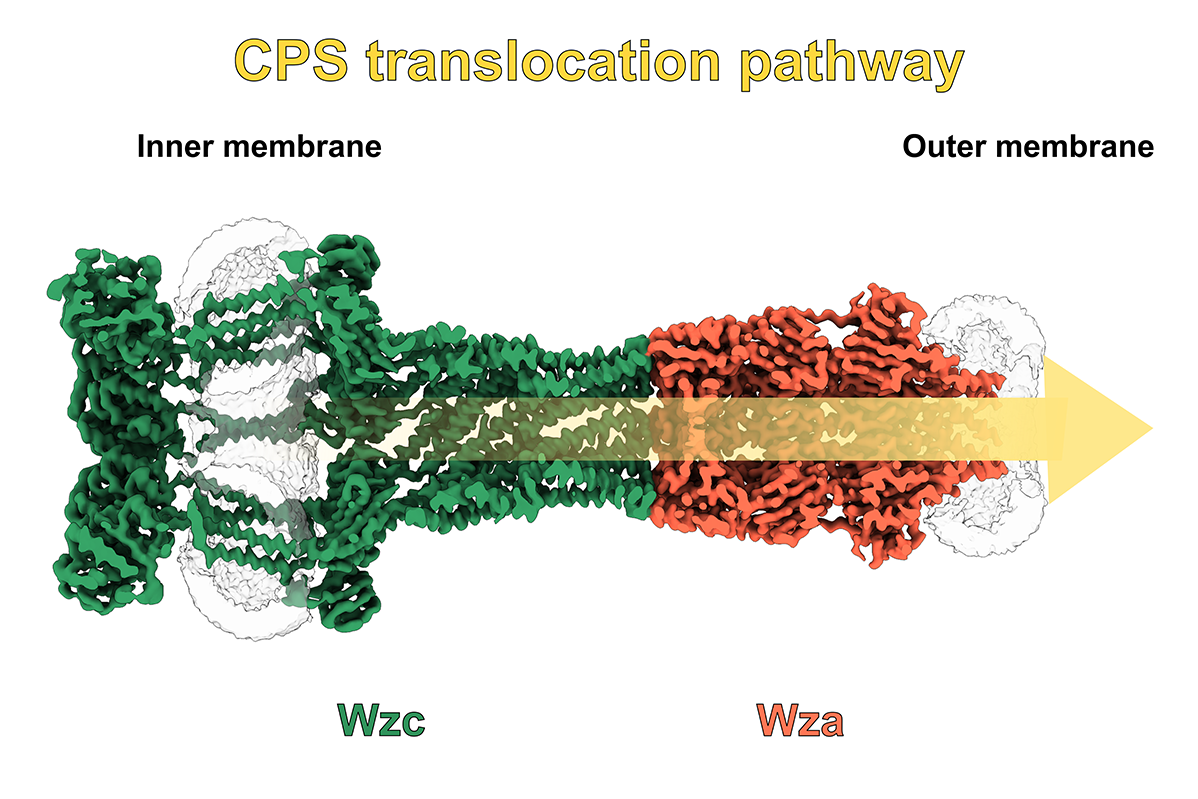
In close collaboration with research groups at the HZI, the current focus of the work lies on novel CRISPR/Cas nucleases, capsid polysaccharide transporters and bacterial natural product megasyntases. Another important goal is the development of innovative pathoblockers that specifically inhibit bacterial virulence factors in order to control infections more effectively and reduce the development of resistance. In addition to synthetic agents and natural products, mini-proteins developed using AI-supported protein design methods are also increasingly being used as potential inhibitors.
Our research
The Department of Molecular Structural Biology is focussing on highresolution structural elucidation of large macromolecular complexes and machines that are important for infection processes. State-of-the-art methods such as single-particle cryo-electron microscopy (cryo-EM) and X-ray crystallography are used for this purpose. The structural biology analyses are complemented by a broad spectrum of molecular biological, biochemical and bio-physical methods.
In close collaboration with research groups at the HZI, the current focus of the work lies on novel CRISPR/Cas nucleases, capsid polysaccharide transporters and bacterial natural product megasyntases. Another important goal is the development of innovative pathoblockers that specifically inhibit bacterial virulence factors in order to control infections more effectively and reduce the development of resistance. In addition to synthetic agents and natural products, mini-proteins developed using AI-supported protein design methods are also increasingly being used as potential inhibitors.
Prof Dirk Heinz
"Seeing is believing" - The beauty of structural biology lies in the fact that the structural information obtained can often be used to directly deduce how the protein molecule in question works.

Dirk Heinz is a German biochemist and structural biologist. He studied chemistry in Freiburg/Germany and received his PhD in structural biology from the University of Basel/Switzerland. After a postdoctoral stay at the University of Oregon and habilitation in Freiburg, he moved to the Helmholtz Centre for Infection Research (HZI) as head of a junior research group and subsequently rose to head of department. In 2011, he took over as Scientific Director of the Center, a position he held until June 2023. Since 2012 he holds a full professorship in structural biology at the Technical University of Braunschweig. In July 2023, Dirk Heinz returned to active research as Head of the Department of Molecular Structural Biology. Dirk Heinz is an EMBO member and holds an honorary doctorate from the University of Würzburg.








Selected Publications
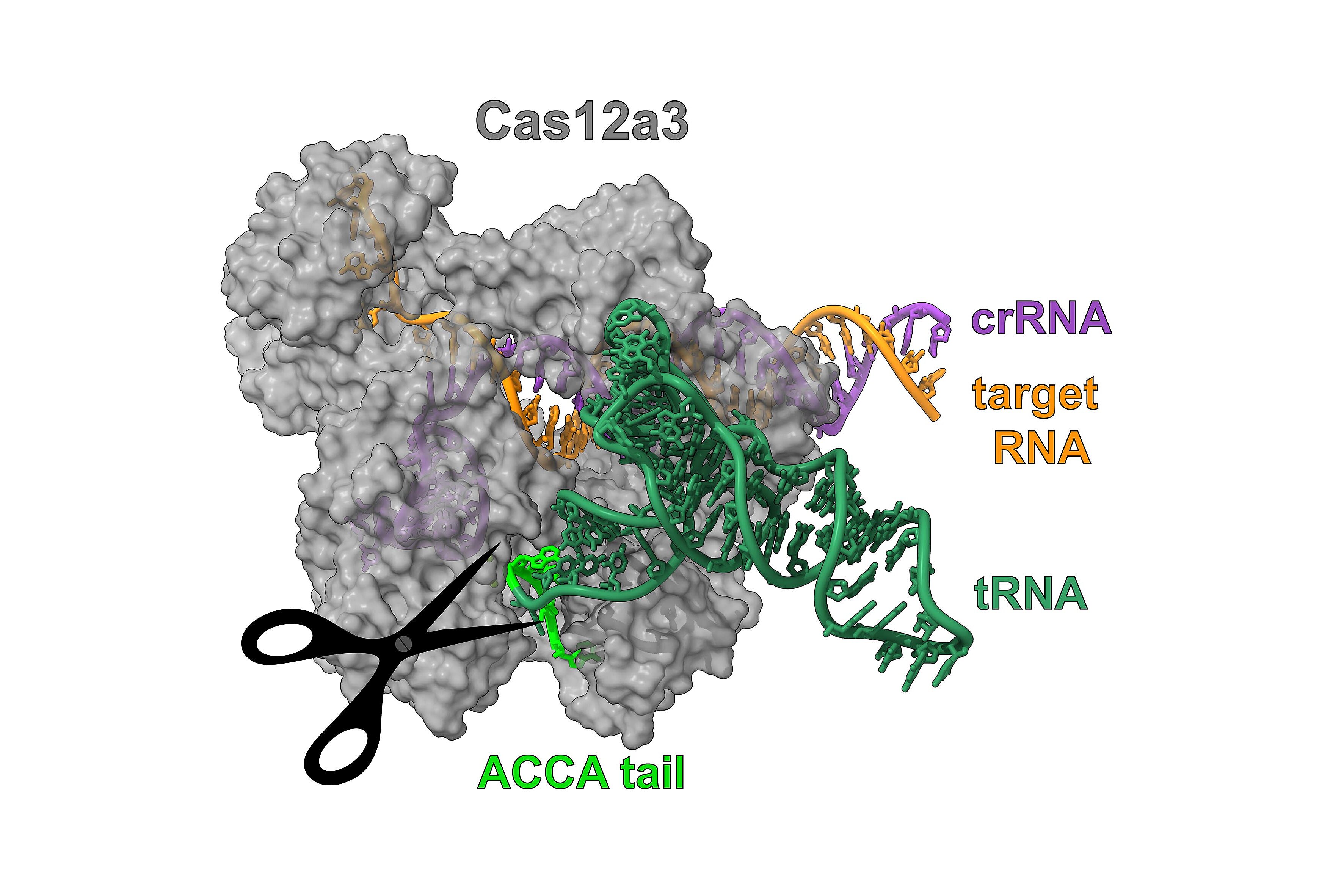
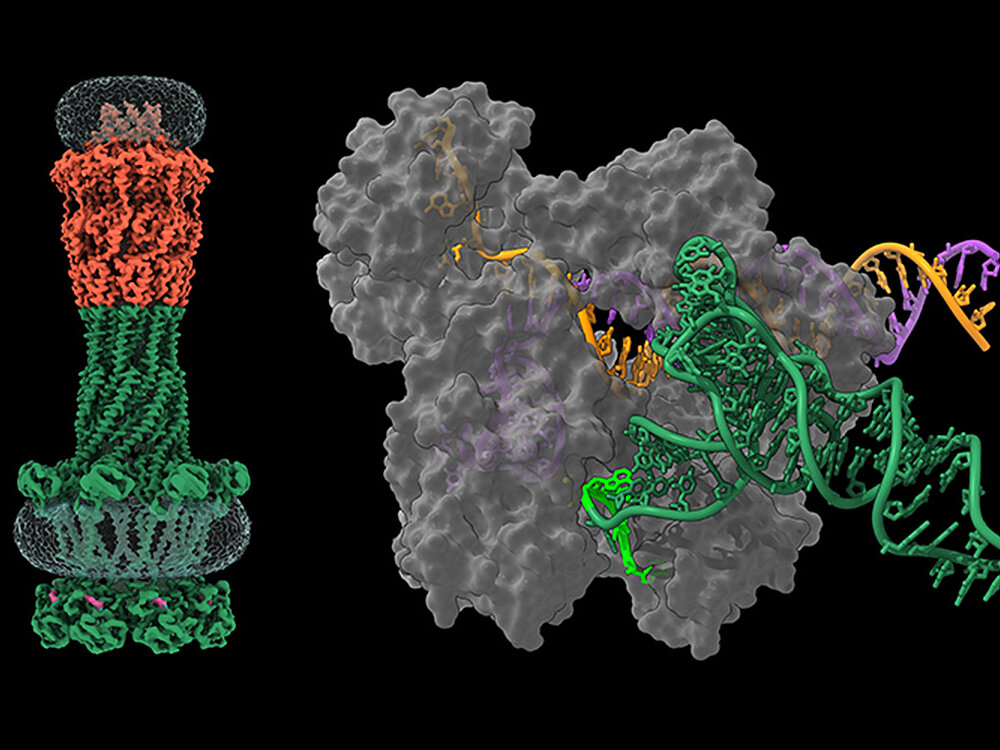
O. Dmytrenko et al., RNA-triggered Cas12a3 cleaves tRNA tails to execute bacterial immuni-ty. Nature, in press (2025).
A. Kling et al., Targeting DnaN for tuberculosis therapy using novel griselimy-cins. Science 348, 1106-1112 (2015).
T. Wollert et al., Extending the host range of Listeria monocytogenes by rational protein de-sign. Cell 129, 891-902 (2007).
H. H. Niemann et al., Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell 130, 235-246 (2007).
W. D. Schubert et al., Structure of internalin, a major invasion protein of Listeria monocyto-genes, in complex with its human receptor E-cadherin. Cell 111, 825-836 (2002).
Publications
An overview of all publications can be found here