The human gut is home to a diverse microbial community. These bacteria play a vital role in maintaining our health by supporting digestion and protecting against disease. They exhibit extreme diversity, with differences not only present at the species level. Even bacteria from the same species with the same genetic composition can look different.
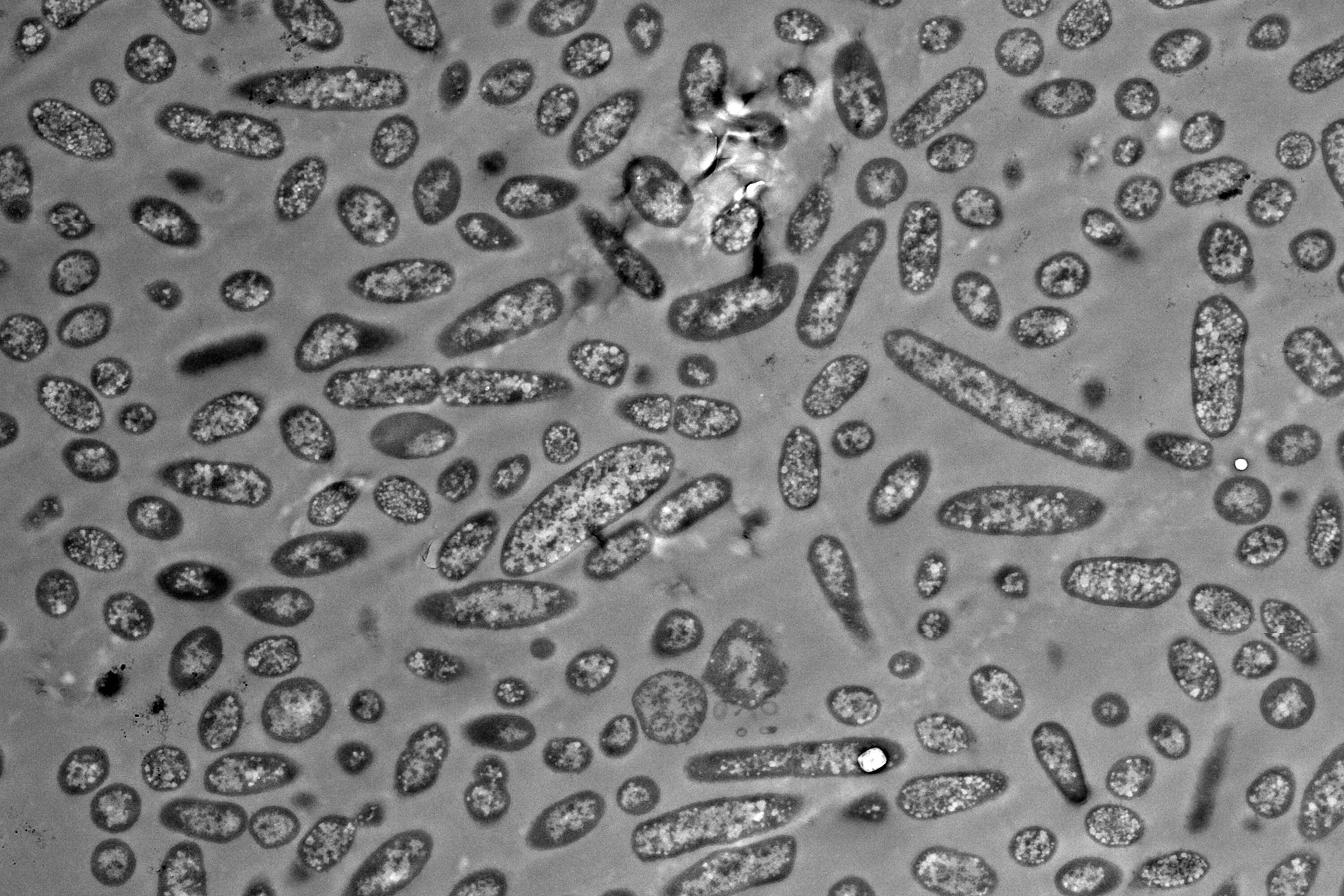
A good example of this is the genus Bacteroides, which represents some of the most common intestinal bacteria and exhibits significant morphological variation. This means they come in different shapes and sizes, known as morphotypes. Until now, the molecular mechanisms behind this diversity and its potential effect on bacterial function have been poorly understood. Researchers from the Helmholtz Institute for RNA-based Infection Research (HIRI), a site of the Braunschweig Helmholtz Centre for Infection Research (HZI) in cooperation with the Julius-Maximilians-Universität Würzburg (JMU), and the Chair of Microbiology at JMU have investigated the functional differences between the various morphotypes of Bacteroides thetaiotaomicron.
Form follows function
The analysis revealed that even within a seemingly homogeneous culture of bacteria, there are significant differences in gene activity between distinct cell morphotypes. “Many of these genes are responsible for producing proteins that are involved in metabolism. This indicates that morphological differentiation is due to metabolic specialization,” explains lead author Alexander Westermann, an affiliated scientist at HIRI and professor at JMU.
“Our results were unexpected,” adds Élise Bornet, a doctoral student in Westermann's lab and first author of the study. “They show that bacterial cell size differences are not just due to the cell cycle. Rather, it appears to be a deliberate strategy of commensals to thrive in an environment as dynamic as the human gut.”
A novel method
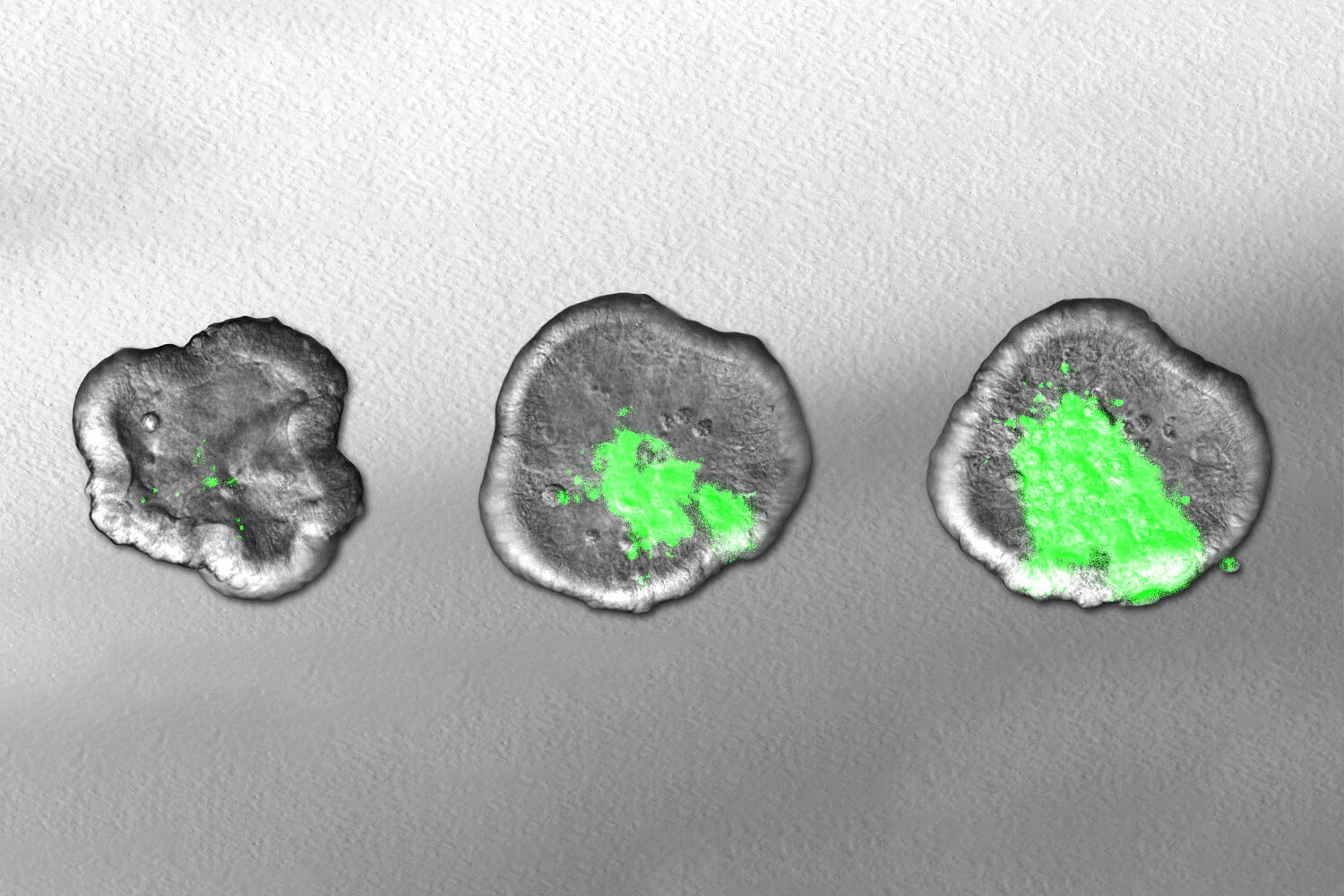
First, the researchers separated and sorted the bacteria based on their size. To characterize the differences in gene activity, the team developed an ultra-sensitive RNA sequencing method in close collaboration with Emmanuel Saliba’s group at the HIRI. This technology makes it possible to determine and analyze the content of ribonucleic acids (RNAs) in biological samples. In addition, the researchers used a staining method called fluorescence in situ hybridization, which allowed them to visualize transcripts directly in the cell. This enabled the team to identify differentially expressed genes as biomarkers for specific cell shapes. The researchers found that these markers are not merely typical of the cell type; some of them are even causally involved in cell size determination.
One advantage of the new method is that it requires only a small amount of material. In fact, just a few cells are sufficient for analysis. “Our protocol is not only important for our current study, but also represents a valuable tool for the research community,” says Westermann. “In the future, our technique could help researchers identify differences in the gene activity of individual cells—both in Bacteroides and in other microbiota species,” adds Bornet.
The gained insights could open up new possibilities for medicine: Since Bacteroides bacteria perform important functions; a better understanding of their diversity could lay the foundation for novel microbiome-based therapies.