Bacteriophages - Viruses that attack bacteria
The enemy of my enemy is my friend - this sentence could also apply to bacteriophages. Bacteriophages (phages for short) are viruses that attack bacteria and are therefore their natural predators. They live up to their name, which is derived from the Greek word for “bacteria-eater”: bacteriophages attach to bacteria, inject their genetic material and reprogram the cells so that they produce large quantities of phage genetic material. Finally, the cells burst and release new phages. These can in turn infect further bacterial cells, as in a chain reaction. In this way, bacteriophages can, for example, “eat” holes in a dense cell lawn with bacteria. However, bacteriophages do not attack bacteria indiscriminately, but are highly specialized in one host species or even only individual strains of this species.
With a size of 30 to 150 nanometers, bacteriophages are extremely small and therefore not visible with a light microscope, but they are very common. No other biological entity (because, strictly speaking, phages are not living organisms) occurs more frequently worldwide. Thus, phages play an important role in all ecosystems. Studies have shown that there are up to ten million bacteriophages in one milliliter of seawater and it is estimated that they kill around one fifth of all bacteria in the oceans every day. This releases nutrients for other species and keeps the ecosystem in balance. Bacteriophages are found in all habitats where their hosts occur. Scientists are therefore looking for new bacteriophages that can fight pathogens, including those commonly found in hospitals, especially in the wastewater of large cities and hospitals.
Phage therapy: opportunities and obstacles
The ability to efficiently decimate bacterial populations makes bacteriophages an attractive therapeutic alternative against infections with multi-resistant germs. Since the discovery of antibiotics, however, phage therapy has been little researched. The increasing spread of antibiotic resistance has brought it back into focus. In phage therapy, the pathogen causing a severe infection is first identified and then one or more suitable phages that are effective against this pathogen are administered. This has already been carried out successfully in individual clinical cases. However, there are high scientific and regulatory hurdles before mass application. For example, an individual formulation must be produced for each infection, which is not feasible under current standards for clinical trials and approvals. To make this possible in the future, the World Health Organization (WHO) and the Global Antimicrobial Resistance Research and Development Hub (Global AMR R&D Hub) are working together to determine the regulatory and legal requirements to support the wider use of phages to combat antimicrobial resistance.
A toolbox for research
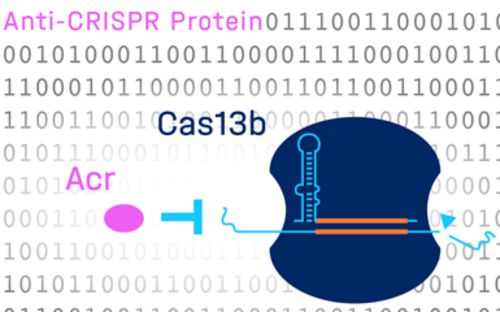
Bacteria are not defenseless against attacks by bacteriophages. Over many millions of years of evolution, bacteria have evolved numerous systems to fend off phage infections. The best-known bacterial immune system is the CRISPR-Cas system, which enables bacteria to recognize and cut DNA segments of bacteriophages - thus preventing infection. Researchers have used this to develop gene scissors that can precisely recognize, cut and edit gene sequences in human cells.
However, there are still many genes in the genomes of bacteriophages whose functions remain completely unknown. These genes could be important sources for the discovery of new molecular tools, which is why researchers at the Helmholtz Centre for Infection Research (HZI) and the Helmholtz Institute for RNA-based Infection Research (HIRI) are elucidating their functions by investigating their mechanisms in bacteriophage-host interactions. Here, a particular focus lies on understanding how bacteriophages take control of their hosts in order to replicate. To this end, the researchers at HZI and HIRI are studying jumbo phages, which carry several hundred genes in their genome, the majority of which is of unknown function. At the HIRI, which focuses on studying the involvement of RNA in infection, scientists also investigate RNA phages. Unlike most phages, which have a DNA genome, the genomes of these phages are particularly small and consist of RNA. To date, RNA phages have received little attention in research. With the introduction of new sequencing technologies, however, this has recently started to change, and it is now possible to detect RNA phages in the environment, which strongly expanded our knowledge about this understudied clade of phages.
Bacteriophages are more than just natural enemies of bacteria: They are central players in almost all ecosystems, a promising source of new biotechnological tools and a beacon of hope in the fight against multiresistant germs. Current research projects at the HZI are deepening our basic understanding of virus-host interactions and providing new biotechnological tools. The next few years will show whether and how these findings will pave the way for widely applicable, safe phage preparations - a decisive step in overcoming the growing threat of antimicrobial resistance.
(cwe)
Status: November 2025
Involved research groups
-
Evolutionary Community Ecology
 Dr Jan Frederik Gogarten
Dr Jan Frederik Gogarten -
Complexes in Phage-infected Cells
 Dr Milan Gerovac
Dr Milan Gerovac -
Molecular Principles of RNA Phages
 Jun Prof Dr Jens Hör
Jun Prof Dr Jens Hör -
RNA Biology of bacterial infections
 Prof Dr Jörg Vogel
Prof Dr Jörg Vogel

![[Translate to English:] Rasterelektronenmikroskopische Aufnahme einer Escherichia coli-Zelle (rot), die von Bakteriophagen (grün) infiziert wird.](/fileadmin/_processed_/1/6/csm_Escherichia_coli_phage_8c7e69405a.jpg)