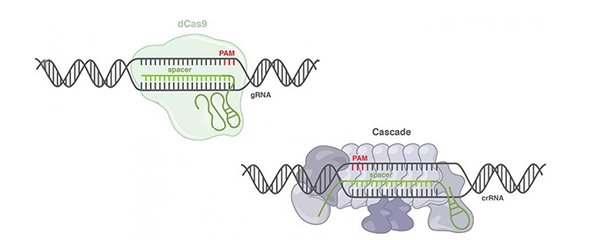
Based on the involved proteins, bacterial CRISPR-Cas systems can be divided into two classes. Class II CRISPR systems require only a single protein for targeted DNA editing. In class I, on the other hand, several proteins must interact in a complex called Cascade. Due to their simple structure, Class II systems have been the focus of research and have so far been the first choice for targeted gene editing. However, they represent only 10% of all naturally occurring systems. “There is a large repertoire of CRISPR-Cas systems that have hardly been investigated and of which we know little about which applications they offer,” says Prof. Chase Beisel, head of the HIRI research group “Synthetic Biology of RNA”. Together with scientists from Duke University and North Carolina State University in the USA, Beisel has programmed a class I CRISPR-Cas system to change gene expression in human cells. The precision of the new system was comparable to the performance of previously established systems. The researchers were able to bind specific DNA sequences and recruit other proteins that allowed the expression of individual genes to be increased or decreased. Such epigenetic modifications are an important factor in gene regulation and in the development of various diseases. The study shows that the complex class I CRISPR-Cas systems are suitable as programmable tools for gene editing. “With class I CRISPR-Cas systems, we are increasing the number of available tools for gene editing and potentially opening up completely new application areas,” says Beisel.
A detailed report by the Duke University can be found here.
Original publication:
Adrian Pickar-Oliver, Joshua B. Black, Mae M. Lewis, Kevin J. Mutchnick, Tyler S. Klann, Kylie A. Gilcrest, Madeleine J. Sitton, Christopher E. Nelson, Alejandro Barrera, Luke C. Bartelt, Timothy E. Reddy, Chase L. Beisel, Rodolphe Barrangou & Charles A. Gersbach: Targeted transcriptional modulation with type I CRISPR–Cas systems in human cells. Nat Biotechnol. 2019 DOI: 10.1038/s41587-019-0235-7