The researchers at the HIRI developed a powerful means to assemble CRISPR arrays, revealing properties to guide CRISPR array design and enabling highly multiplexed targeting. Employing this new approach revealed new insights into the biology of CRISPR-Cas systems, the source of CRISPR technologies.
What is it all about?
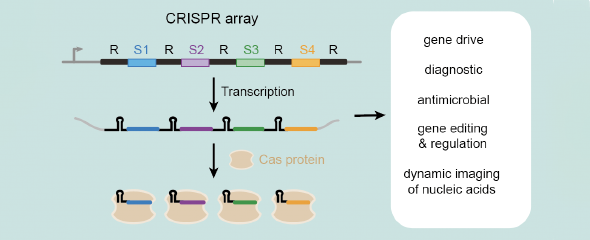
CRISPR technologies represent powerful tools for genome editing that are revolutionizing basic research, medicine, biotechnology, and agriculture. One key feature of these technologies is their ability to multiplex—or target multiple genes at one time—by using multiple guide RNAs. One major challenge is how to best encode these guide RNAs. The exquisite solution CRISPR-Cas systems offer is in the form of CRISPR arrays. These arrays consist of identical sequence repeats alternating with unique spacers. Each repeat-spacer pair becomes a guide RNA. CRISPR arrays are highly compact and can encode hundreds of unique guide RNAs. However, these same arrays have proven incredibly difficult to construct, limiting our basic understanding of their properties and how they can be used for multiplexing applications with CRISPR technologies.
What are the key findings?
In Nature Communications, Beisel and his postdoctoral researcher Dr Chunyu Liao report a powerful means to construct individual CRISPR arrays. This approach, which they call CRATES (for CRISPR Assembly through Trimmed Ends of Spacers), allowed them to create not only long CRISPR arrays but also libraries of CRISPR arrays, enabling highly multiplexed editing or rapidly screening combinations of edits. They further used the arrays to gain new insights into the biology of CRISPR-Cas systems, working toward a more advanced understanding of this source of CRISPR technologies.
Why is this new method so powerful?
CRATES has the potential to advance the many applications of CRISPR technologies requiring multiplexing, such as editing multiple genes to simulate complex diseases, rapidly optimizing industrial bacteria for sustainable chemical production, improving CRISPR gene drives, and developing multi-input diagnostics. Furthermore, the researchers were able to derive important insights into CRISPR arrays that represent one-half of all CRISPR-Cas systems yet have been far less studied or applied.
For further reading, please see the "Behind the paper" article on nature.com
Original publication:
Chunyu Liao, Fani Ttofali, Rebecca A. Slotkowski, Steven R. Denny, Taylor D. Cecil, Ryan T. Leenay, Albert J. Keung & Chase L. Beisel: Modular one-pot assembly of CRISPR arrays enables library generation and reveals factors influencing crRNA biogenesis. Nat Commun 2019 DOI: 10.1038/s41467-019-10747-3